ridgeFit#
Purpose#
Fit a linear model with an L2 penalty.
Format#
- mdl = ridgeFit(y, X, lambda)#
- Parameters:
y (Nx1 vector) – The target, or dependent variable.
X (NxP matrix) – The model features, or independent variables.
lambda (Scalar, or Kx1 vector) – The L2 penalty parameter(s).
- Returns:
mdl (struct) –
An instance of a
ridgeModelstructure. An instance named mdl will have the following members:mdl.alpha_hat
(1 x nlambdas vector) The estimated value for the intercept for each provided lambda.
mdl.beta_hat
(P x nlambdas matrix) The estimated parameter values for each provided lambda.
mdl.mse_train
(nlambdas x 1 vector) The mean squared error for each set of parameters, computed on the training set.
mdl.lambda
(nlambdas x 1 vector) The lambda values used in the estimation.
Examples#
Example 1: Basic Estimation and Prediction#
new;
library gml;
/*
** Load data
*/
// Specify dataset with full path
fname = getGAUSSHome("pkgs/gml/examples/qsar_fish_toxicity.csv");
// Split data into training sets without shuffling
shuffle = "False";
{ y_train, y_test, X_train, X_test } = trainTestSplit(fname, "LC50 ~ . ", 0.7, shuffle);
// Declare 'mdl' to be an instance of a
// ridgeModel structure to hold the estimation results
struct ridgeModel mdl;
// Set lambda vector
lambda = seqm(90, 0.8, 60);
// Estimate the model with default settings
mdl = ridgeFit(y_train, X_train, lambda);
After the above code, mdl.beta_hat will be a \(6 \times 60\) matrix, where each column contains the estimates for a different lambda value. The graph below shows the path of the parameter values as the value of lambda changes.
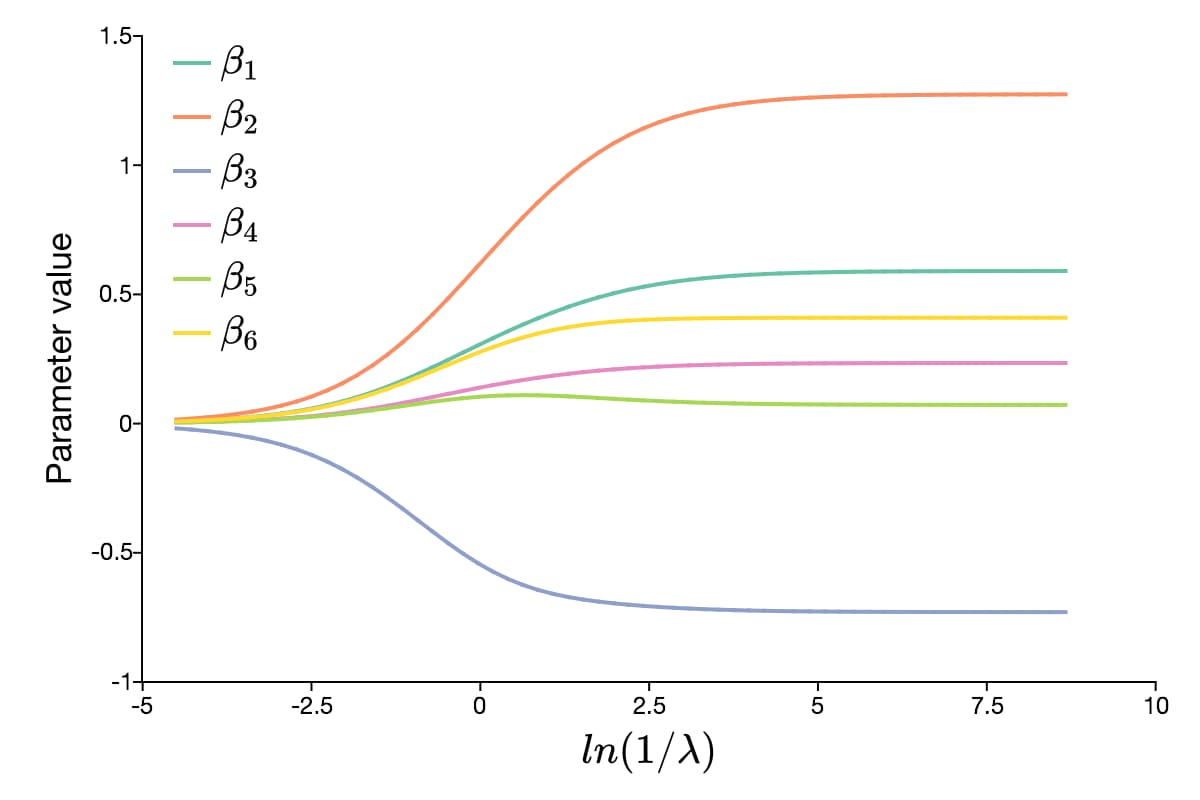
Continuing with our example, we can make test predictions like this:
/*
** Prediction for test data
*/
y_hat = lmPredict(mdl, X_test);
After the above code, y_hat will be a matrix with the same number of observations as y_test. However, it will have one column for each value of lambda used in the estimation.
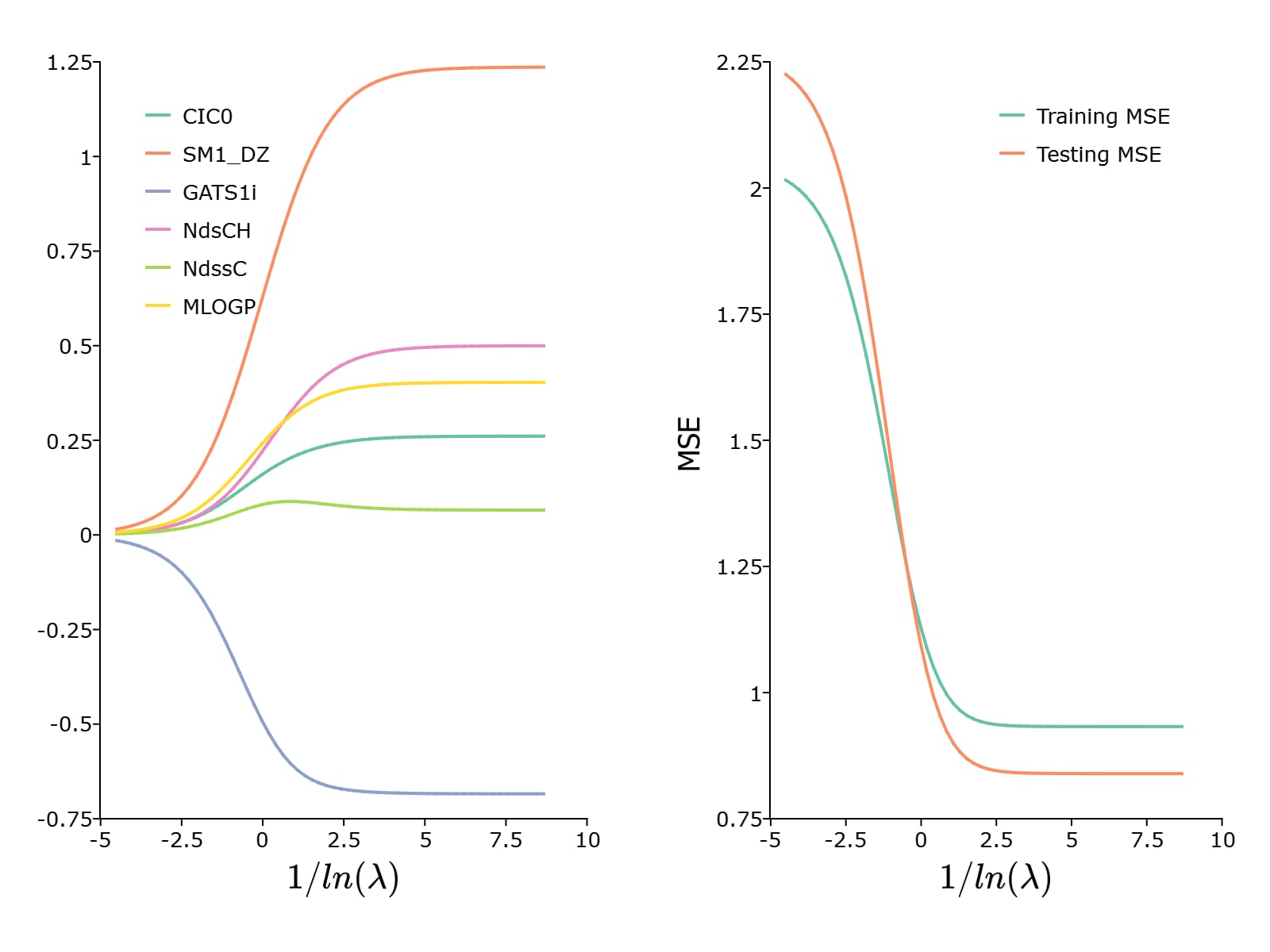
To plot the paths of the coefficients and the MSE, we can use the plotLR() function
test_mse = meanSquaredError(y_test, y_hat);
/*
** Plot results
*/
plotLR(mdl, test_mse);
This results in the following plot:
Remarks#
Each variable (column of X) is centered to have a mean of 0 and scaled to have unit length, (i.e. the vector 2-norm of each column of X is equal to 1).
See also
